Providing Reliable, Reproducible and Valid Results with Bioinformatic Versioning¶
BugSeq helps many clinical and public health labs to quickly identify pathogens, outbreaks, antimicrobial resistance and more. In order for labs to utilize BugSeq data, they need evidence that the results are both correct and reproducible. Imagine if a system diagnosed the same patient with different conditions depending on what day of the week it was - that system would not be very useful. More concretely, given a set of inputs, a system must produce the same outputs to be useful in a high-stakes setting. Our analysis solutions are no different - labs trust us not only for our accuracy, but also for our reproducibility. It allows them to rely on our results, and many customers have used this property to validate LDTs based on BugSeq. So how do we do it?
When a user submits data to BugSeq, many servers turn on and start crunching through the data. On those servers, we run hundreds of tools that access many different databases. For example, a given metagenomic analysis may utilize more than eight different databases within an hour runtime (more on that in a future post).
What is a Version¶
First, it’s important to understand how software and databases are typically versioned. Then we can review how BugSeq uses those versions to provide reproducible, versioned analysis.
Software Versioning¶
Git References¶
For a given tool, versioning can mean different things. Typically, the code itself is versioned. You’ll see this in the form of git references. Think of this version as a unique identifier that represents all the lines of code. An example is 7198b7da2a5a244352204bb5c6bc68a4d50e08ce.
Git references are nice because they specify the actual code that runs. However, git references are plentiful and not semantically meaningful. It is quite difficult to understand what is new in one reference versus another by looking at it.
Furthermore, the code version only tells part of the story. The same code may run differently depending on where it is run. For example, you may get different results running on Mac versus Linux servers, with Internet connection versus offline, and more.
Releases¶
To make git references more manageable, many tools have turned to versioned releases (e.g. v2.5.1). These typically represent both code and built artifacts. Tools will also typically release a changelog to help understand what’s new in a specific version. After time has passed, it becomes increasingly difficult to build and run old code. Having a pre-built application means you can just run it, the same way you did before.
Here is an example release for minimap2:
Containers/Images¶
Even with a pre-built application, behavior can vary depending on the environment it runs in. As an example, many legacy python2 applications will not run in a modern python environment.
Containers are running environments that capture both an application and its dependencies. They are also portable between servers, and easy to run years later. Containers are each uniquely identified by a long identifier.
Database Versioning¶
Some tools leverage databases to produce results. As an example, think of an alignment tool that must align reads against a database. Even if the software version stays the same, using a different database will produce different results. Many tools even update their database each time they run, which makes reproducibility very difficult.
Databases can be versioned just like software. Typically, database versions are denoted by dates rather than increasing numbers because they are easier to reason about.
Pipeline Versioning¶
A pipeline is typically a combination of many tools, databases, and the logic that flows data through the tools. In effect, you can think of a pipeline version as a combination of many other software and database versions.
Keeping the same pipeline running reliably for years can be very challenging. Imagine trying to investigate a finding from last year, only to find that one of many tools no longer builds, or one of many databases is no longer available.
Locking Down Everything¶
BugSeq offloads the complexity of versioning so you can just pick a version, and have confidence it will run the same way it did last time - no need to worry about software versions, database versions, building tools, or the runtime environment surrounding them.
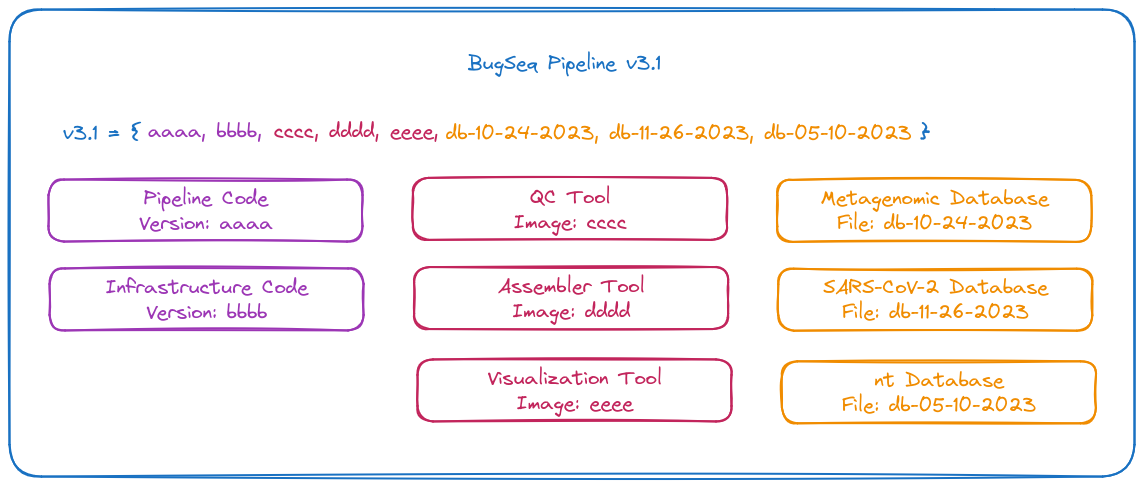
So what’s in a BugSeq version? It is a combination of all our code, tools and databases that can be run in exactly the same way, any time, to produce the same results:
When we release a version, we save all the code, images and databases. We then store references to all of those as a single “version”, and provide a name.
These are the versions you can select for each analysis you run.
Smoothing the Upgrade Process¶
While reproducibility is nice, so is having the latest and greatest software. Typically, upgrading requires that you validate that the new version produces the same or better results as the old.
BugSeq can reprocess old data on newer versions to give a side-by-side comparison on the same data, yielding much easier upgrades. This allows customers to get the best of both worlds - reproducibility for day-to-day operations, and easy upgrades when you are ready.
Summary¶
BugSeq enables users to select which version analysis to run their data through. A version consists of code, tools and databases; beyond labels and numbers, versions enable you to get reproducible results year after year.
Want a solution where reproducibility and upgrades are already solved? To learn more about how BugSeq can help your lab: