BugSeq Bioinformatics Inc. Collaborative Effort to Develop and Validate an end-to-end Agnostic Diagnostic Assay Published¶
Introduction¶
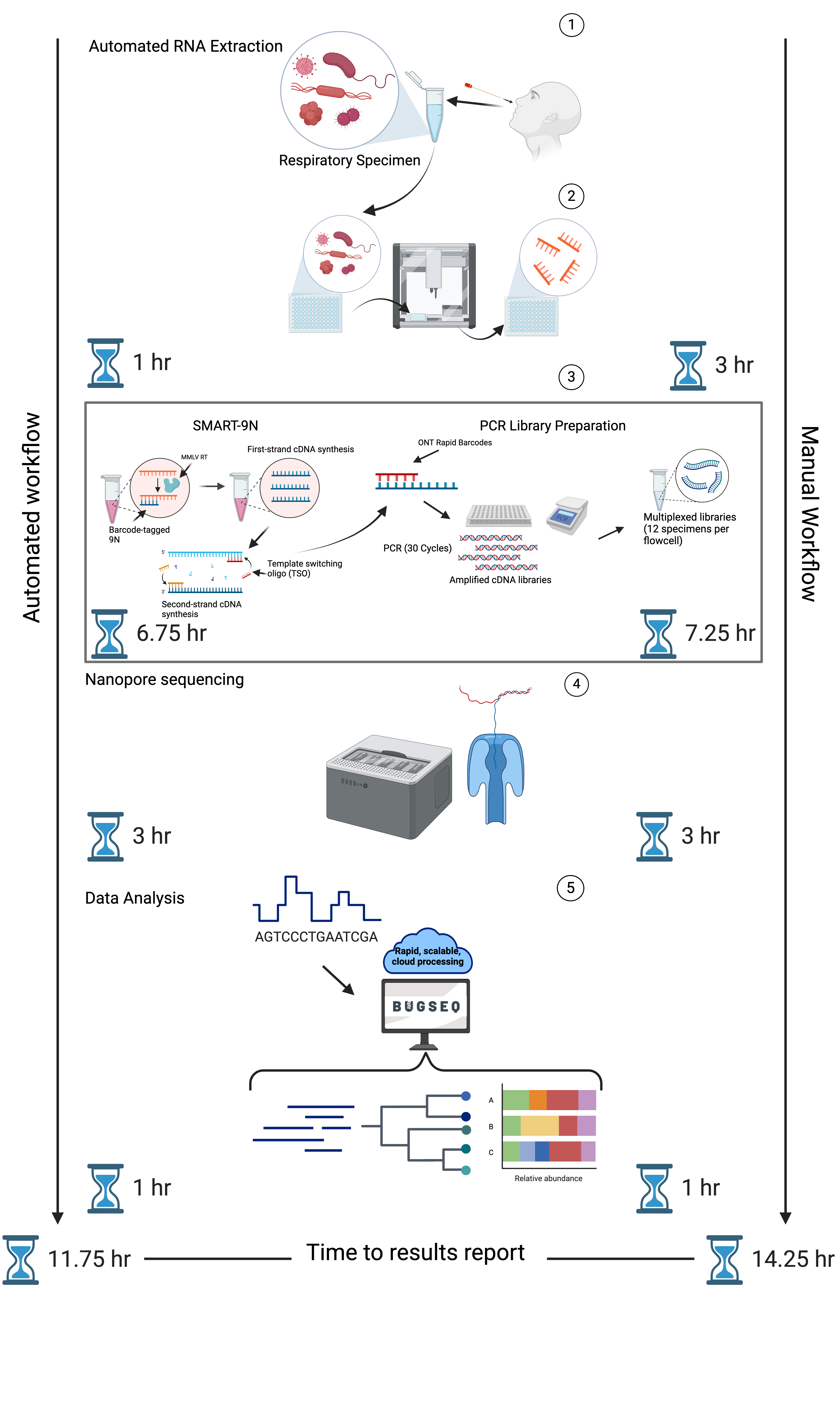
BugSeq partnered with University of British Columbia Researchers, along with clinical and public health collaborators at Vancouver Coastal Health, BC Centre for Disease Control, and Canada’s Michael Smith Genome Sciences Centre to develop, automate, and validate an end-to-end metagenomic sequencing assay for agnostic detection of respiratory viral pathogens within 12-hours. The work was funded by the Biomedical Advanced Research and Development Authority (BARDA), part of the Administration for Strategic Preparedness and Response within the U.S. Department of Health and Human Services under contract number 75A50122C00027.
The results of this collaborative project were published last week in The Journal of Infectious Diseases. Gauthier et al. outlined the development and validation of a metagenomic sequencing assay, termed Rapid Pathogen Identification through Metagenomic Next-Generation Sequencing (RAPID-mNGS).
What did the authors find?¶
Through laboratory and bioinformatic optimization and automation, multiplexing of up to 55 specimens was possible using an Oxford Nanopore GridION sequencing device, providing sample-to-answer results within 12-hours.
The authors overcame many translational barriers to mNGS adoption in clinical settings, including reductions in turnaround time, cost, and automation to reduce hands-on time. Analytical and clinical validation of the RAPID-mNGS assay revealed that the method is precise, yields low limits of detection (103-104 copies/mL), is highly specific (100%), and sensitive (86.8%; RT-PCR Ct < 30). Clinical validation of the RAPID-mNGS assay on specimens collected from Vancouver Coastal Health and the BC Centre for Disease Control revealed the presence of other viral pathogens missed by conventional testing, which were confirmed through a reference standard multiplex PCR assay.
Gauthier et al:
“The incidental detection of viral pathogens in “negative” specimens emphasizes the potential utility of an agnostic diagnostic assay in clinical practice.”
BugSeq reported optimizations in our bioinformatic workflow, which led to consistent reporting of interpretable results reports in less than 1-hour. A novel pathogen exercise using databases that had certain viruses masked revealed that the BugSeq metagenomic classification pipeline can detect novel pathogens with high accuracy.
Gauthier et al:
“Our team successfully identified the viral family for the influenza samples (Orthomyxoviridae) and the genus for the RSV samples (Orthopneumovirus), without overclassifying to other Orthomyxoviridae or Orthopneumovirus species.”
Why is this important?¶
Existing molecular diagnostic techniques, while cost-effective and actionable, are limited in their ability to detect novel pathogens. Metagenomic sequencing offers a pathogen-agnostic diagnostic approach, which is an invaluable attribute in the context of pandemic preparedness and diagnosis of infection by rare etiologic agents. While there has been previous work performed to demonstrate the usefulness of mNGS as a proof-of-concept in clinical settings, translational barriers, such as high-cost, technical workflows, lengthy turnaround times, poor sensitivity, and difficult to interpret results have hindered widespread adoption of mNGS in clinical practice.
Through the partnership summarized in this publication, BugSeq, alongside our academic and clinical collaborators developed a rapid, seamless, end-to-end assay, which provides interpretable and user-friendly results reports to overcome many of these translational barriers. While there are some remaining challenges that need to be overcome to drive this technology towards broader clinical use, the results of this partnership represent a strong step towards the realization of NGS-based infectious disease diagnostics.
Interested in learning more?¶
- Contact Us: contact@bugseq.com
- Request a Quote: bugseq.com/quote
- Publications: bugseq.com/publications