Pipeline Change Log¶
Latest¶
Known Issues¶
- Mean Phred scores per base in summary report are calculated as a simple average of all base Phred scores, instead of incorporating considerations for a logarithmic scale. Additional details are available here.
- AMR genes may be reported on a plasmid for drugs which are not included in the predicted phenotype panel.
- E. coli shiga toxin detection may miss multiple alleles if both occur in the same sample and data was generated with a Nanopore or PacBio sequencer.
Added¶
- Reporting of text warnings at the top of reports for anomalous data. The first warning implemented is triggered for RNA viruses detected in DNA samples.
Fixed¶
- Assembly completeness coloring is fixed to more logical colors (see MultiQC Issue #2450).
- Running the same sample through BugSeq multiple times on the same analysis version rarely resulted in different plasmids detected. The issue was reported by BugSeq and a fix implemented in collaboration with the upstream tool maintainers.
- Remove the hidden average and median read length columns in the General Statistics table of summary reports. Users previously could enable visualization of these columns, which would show inaccurate data; inaccuracy was the result of how MultiQC calculated them.
Changed¶
- AMR database update.
- Improved AMR prediction for fosfomycin in Klebsiella pneumoniae by selective reporting of fosA alleles.
- Improved aminoglycoside AMR prediction.
- Report 10 instead of 5 top taxa in the metagenomic classificaiton plots in the summary report
- Better protein knockout detection for AMR prediction.
- Read classification CSV outputs classification of host range of plasmid, regardless if the sample is a bacterial isolate or not. This change ensures the read classification CSV matches results from the krona plot and reports.
- Improved reporting of viruses by using their common names when appropriate. For example, Influenza A virus will be used in many places instead of Alphainfluenzavirus influenzae. See the NCBI Insights post for details.
- Improved annotation of blaSHV alleles from Tsang et al.
- Improved prediction of nitrofurantoin resistance in E. coli based on Dulyayangkul et al.
- Improved prediction of carbapenem resistance in Pseudomonas aeruginosa by curating public mutations.
- Improved prediction of vancomycin resistance in Enterococcus species by requiring select combinations of van operon genes. See Coll et al. for details.
- Improved Nanopore host filter by using dynamic filtering criteria based on alignment length.
- Improved prediction of beta-lactam resistance in Acinetobacter calcoaceticus/baumannii complex by incorporating information on intrinsic beta-lactamases and insertion sequence detection into prediction.
- BugRef (curated reference sequence) database updated. Fungal representation is improved including manually curated Trichophyton genomes from the T. mentagrophytes complex.
- Improved which drugs are reported for agents of bioterrorism to match CLSI M45-A3.
- SARS-CoV-2 lineage database update.
- Mycobacterium tuberculosis antimicrobial resistance database update to WHO v2. Pretomanid resistance prediction is newly reported.
5.3 - May 17, 2024¶
Known Issues¶
- Mean Phred scores per base in summary report are calculated as a simple average of all base Phred scores, instead of incorporating considerations for a logarithmic scale. Additional details are available here.
- AMR genes may be reported on a plasmid for drugs which are not included in the predicted phenotype panel.
- E. coli shiga toxin detection may miss multiple alleles if both occur in the same sample.
- Running the same sample through BugSeq multiple times on the same analysis version may rarely result in different plasmids detected. A fix has been submitted to the upstream tool.
Median Read LengthandAverage Read Lengthcolumns (Group:Read Quality Control) reported in hidden columns in the General Statistics table of the summary report are inaccurate. These columns must be explicitly unhidden viaConfigure columnsto view the data. This error results from MultiQC inaccurately calculating these statistics from FastQC outputs. Please use theMedian Read Lengthcolumn shown by default, which is accurately calculated by a custom BugSeq tool.
Added¶
- Antifungal resistance prediction for Candida auris.
- New columns and explanations to the plasmid tables in per-sample and summary reports:
- Subtype: plasmid subtype cluster IDs can be used to identify near identical plasmids and are more granular than cluster IDs.
- Predicted mobility: mobility prediction is based on the presence of relaxase, mate-pair formation and oriT sequences. A plasmid is classified as conjugative if it contains at least a relaxase and a mate-pair formation marker. Plasmids containing either a relaxase or an oriT but are missing the mate-pair formation marker are classified as mobilizable, while plasmids that are missing a relaxase and an oriT are classified as non-mobilizable.
- Mash distance to nearest NCBI reference sequence: see work by Robertson et al. for details.
- Detection and subtyping of C. botulinum toxin in appropriate Clostridium species.
- E. coli serotyping and shiga toxin detection.
- Report de novo assembly sequencing depth in the summary report for bacterial isolates. Note that this is slightly different than reference-based sequencing depth, which uses a species-specific reference genome to calculate coverage.
- Legionella pneumophila Sequence Based Typing (SBT).
- Streptococcus pneumoniae serotyping.
- Report multiple MLST schemes for organisms where there are multiple accepted schemes. If your organism of interest has multiple schemes but one or more are not reported, contact us to have it added.
- Output VCF files from Illumina analyses of viruses.
- Minority variant calling for M. tuberculosis resistance from Illumina data.
- Shigella genotyping.
- Report sequencing depth of genomes in the per-sample reports for Nanopore metagenomic samples.
- Report read duplication (column hidden by default) and remove duplicates for Illumina bacterial isolates.
Fixed¶
- Plasmid host range in the summary report plasmid overview table was previously blank.
- Cluster code generation which previously may have reported a close cluster code for genomes which were in fact distant.
Warning
This is a breaking change for cluster code addresses, as the address of old isolates may be updated upon a new analysis. BugSeq has contacted users reliant on cluster codes given the significance of this change.
- Bug affecting classification of contigs to the genus rank or above. Contigs originating from novel or under-represented species may have been unclassified. Benchmarking results and species-rank classifications are unchanged by this fix.
.fnaextension may have been shown in assembly completeness plots of per-sample reports.- Reference-based coverage calculation (in per-sample reports) was calculated based on PCR duplicate-marked alignments. However, duplicate marking was overly aggressive and coverage may have been underestimated.
- Improved detection of AMR genes that were broken at the ends of contigs. This resulted from gene annotation tools (e.g. Prokka, Bakta) disabling gene calling at the ends of contigs.
Changed¶
- Improved accuracy for plasmid detection by using a curated repetitive sequence filter during plasmid identification. Previously, there may have been false negative detection of an IncX3 plasmid.
- Improved accuracy for plasmid detection by curating the plasmid reference sequence database. Excluded <0.02% sequences which are likely chromosomal.
- Revert: “Improved use of assembly graph for classifying Illumina contigs” in v5.2. In v5.2, there was an assumption that errors in the Illumina assembly graph were rare and the taxonomic bin could be assumed to be the same for an entire subgraph within the graph. After identifying several cases where graph edges connected nodes from unrelated taxa, we have reverted this change. BugSeq now uses a probablistic framework to assess the edges between nodes in the assembly graph. This framework incorprates assembly graph edges, paired-read edges (if available) and neighbor labels. Subgraphs are no longer assumed to reflect the same taxon. Accuracy is improved over v5.2:
| v5.2 | Latest |
|---|---|
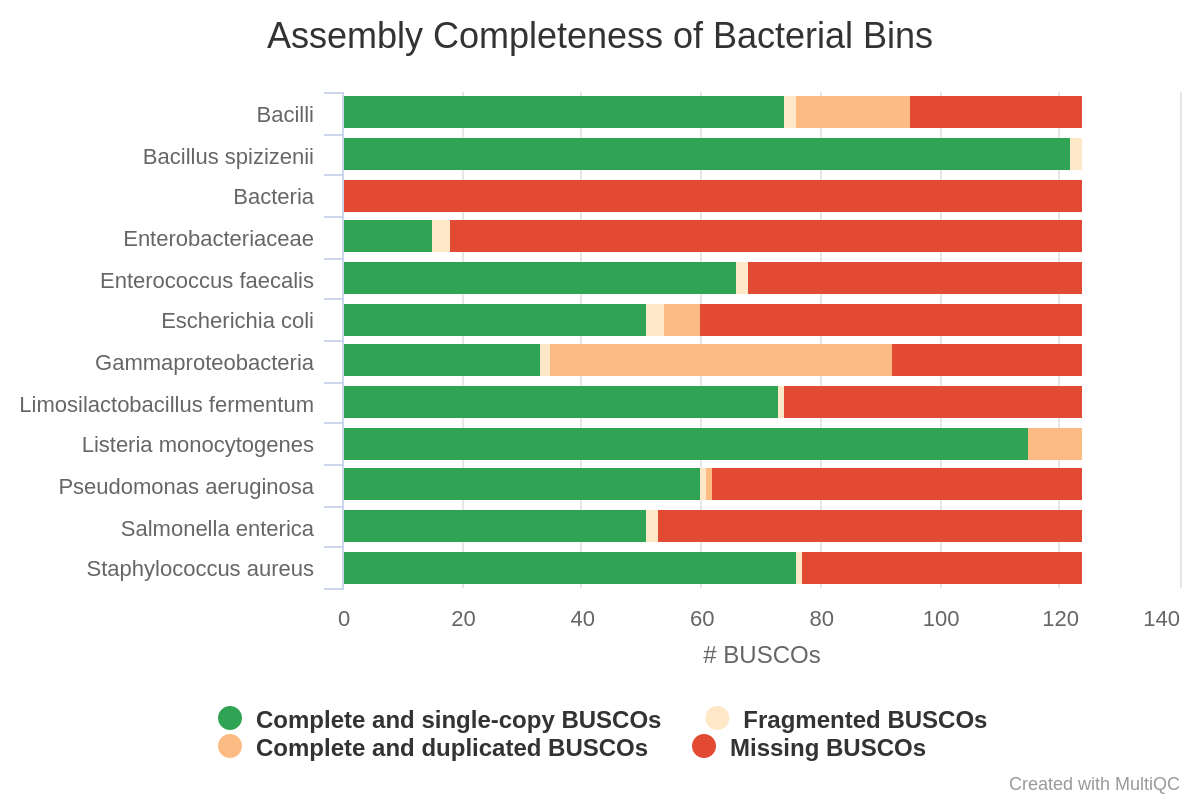 | 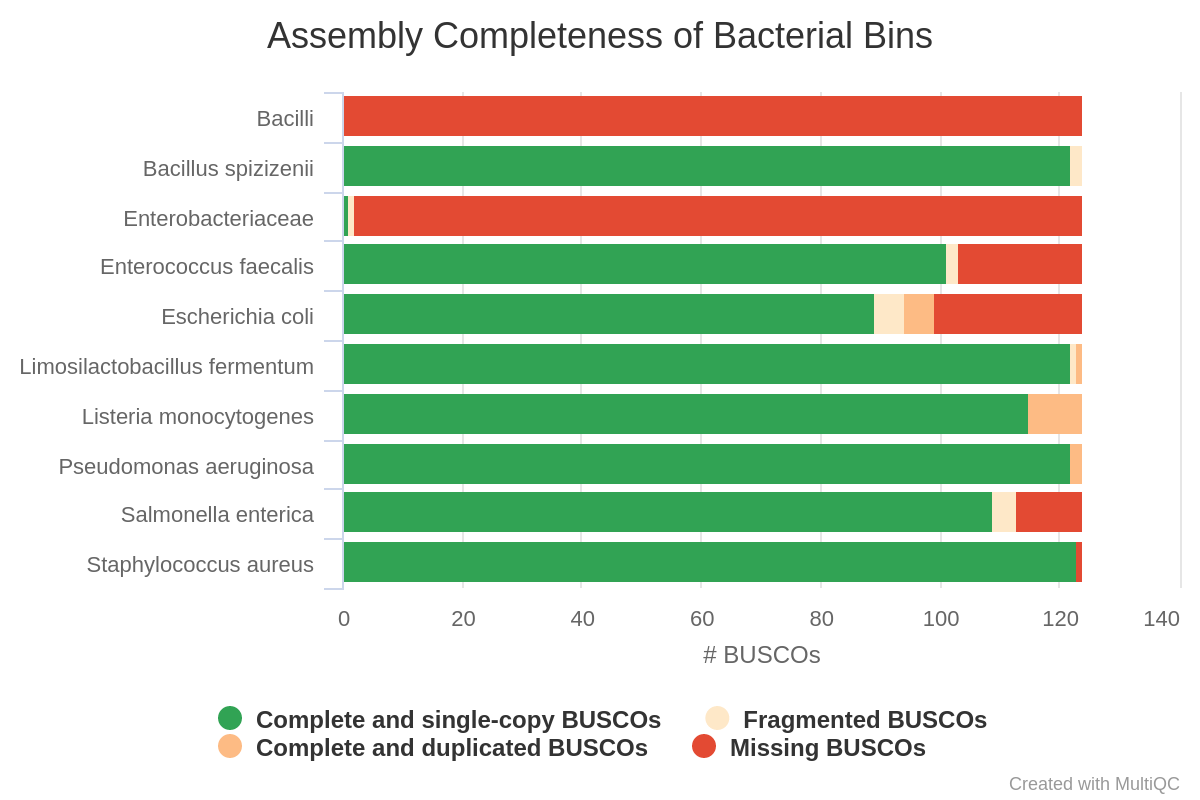 |
- Improved read trimming for Illumina and now enabled read trimming for other sequencing platforms. The improvement handles cases where there are very low quality bases at the ends of reads.
- Faster Nanopore assembly by using the new
lr:hqminimap2 preset for read-to-assembly alignment when appropriate. - Rename BugSeq Default DB to BugRef Curate DB in reports. See recent publication from our team on curation methods.
- FAST5 files are no longer accepted for upload. We recommend using ONT’s dorado for basecalling.
- Outbreak analysis
Distance MatrixandCluster Addresseshave been combined as separate sheets in a single excel output per-organism. A third sheet has been added with a distance matrix including only the samples of the viewed analysis. - Disambiguate host and vector/control sequences in the summary report General Statistics table. This change enables users to better understand why reads were filtered from the sample before metagenomic classification.
- Improved visualization of refMLST outbreak clusters.
- Improved recovery of contigs from repetitive regions when assembling short reads. The minimum contig length of short read assemblies has been decreased from 1000bp to a dynamically set threshold based on input read length.
- Further annotation of OXA-family beta-lactamases.
- Improved classification of Clostridium haemolyticum and Clostridium novyi.
- Updated reference sequence database. This update bring prophage masking, which should yield improved classificaitons for reads deriving from phages. It also introduces additional representation of Influenza A and Morganella species for improved detection.
- Classify more Illumina reads to deeper ranks by performing a two step classification including a profiling step and then a binning step.
- Classify viruses to more commonly known names, such as Influenza A virus instead of Alphainfluenzavirus influenzae.
- Improved host filtering of Nanopore and PacBio reads by incorporation of base quality into the filtering algorithm. Host filtering is more specific because it will not filter reads which are divergent from human yet accurate.
- Dynaminc minimum alignment length for Nanopore read-based classification. This change improves specificity; there should now be fewer false positive species detected at very low abundance.
- Rotate circular contigs for better detection of AMR genes at the ends of contigs.
- Update
pangolin-datato 1.27.
5.2 - Feb 8, 2024¶
Known Issues¶
- Mean Phred scores per base in summary report are calculated as a simple average of all base Phred scores, instead of incorporating considerations for a logarithmic scale. Additional details are available here.
- AMR genes may be reported on a plasmid for drugs which are not included in the predicted phenotype panel.
Added¶
- Influenza analysis enabling typing of Influenza A using amplicon or metagenomic approaches.
- Quality control note to accompany quality control flag of Pangolin SARS-CoV-2 variant calling (see MultiQC PR #2157)
- Hepatitis B virus genotyping reported on summary and per-sample reports from nanopore amplicon sequencing data.
- Output assemblies have contig length and plasmid name (if they are classified as a plasmid) reported in sequence headers. The reported format is:
>contig1 len=4
AGTC
>contig2 len=4 plasmid=AA800
CTGA
- Illumina barcode crosstalk correction based on a method we described in Nanopore metagenomic sequencing for detection and characterization of SARS-CoV-2 in clinical samples.
Warning
Barcode crosstalk correction for Illumina is based on a general approximation of the crosstalk rate seen across Illumina datasets. The observed barcode crosstalk rate depends on many factors, such as the use of dual-indexed versus single-indexed adapters. For customization to your experimental design, get in touch.
Required data for application of crosstalk correction
Barcode crosstalk correction for Illumina relies on detection of the sequencing run ID in read headers of the FASTQ files. If the sequencing run ID is not found (eg. files downloaded from SRA), crosstalk correction will not be applied.
- Confidence reporting for the absence of antimicrobial resistance. As detailed in reports, high confidence reflects that BugSeq obtained a complete or near complete genome for the organism and therefore judges it unlikely that there are factors predicting AMR which were missed. Conversely, incomplete genomes have lower confidence because predictors of resistance may have been missed.
- Visualization of refMLST outbreak analysis results with a minimum spanning tree.
- Generate consensus sequences for all viruses from Illumina sequencing data.
- Hidden columns to General Statistics table of per-sample report with additional details on each taxon (eg. rank and NCBI Taxonomy identifier).
Changed¶
- Minimum contig length for DNA analyses is set to 1000bp and for RNA/TNA analyses is dynamically set based on read length. This change significantly improves assembly quality and binning.
- New ensemble metagenomic classifier incorporating the results of multiple metagenomic classifiers. This is our largest upgrade for metagenomics since Version 2.0. Performance is improved for low abundance pathogens while maintaining leading precision. Notably, there are no longer results for both the read-based and assembly-based classifiers, simplifying outputs and interpretation of results.
- Improved nanopore RNA assembly accuracy. This change improves the detection of known and novel viruses using nanopore RNA-Seq.
- Update pangolin database for SARS-CoV-2 lineage calling.
- Plasmid database update which improves host range prediction.
- Update curated reference sequence database, bringing:
- Improved representation of eukaryotes, including fungi and protozoa.
- Improved filtration of genomes which are taxonomically misidentified.
- Hide Hepatitis B virus variants which derive from genotypic variation.
- Improved quality control of Legionella serogrouping if serogroup 14 is detected, which now reports a QC failure.
- Output assemblies are sorted from largest to smallest contig.
- Polishing of short read isolate assemblies resulting in higher base accuracy.
Note
Allele distances from refMLST may be reduced based on this change.
- BugSeq no longer filters reads from animal species (eg. mouse, rat, pig, etc.) before processing by default. These genomes were found to be contaminated with microbial sequences and filtering impacted downstream analyses. Human reads continue to be filtered by default.
Processing metagenomic data from animals
If you are performing metagenomic sequencing of an animal, get in touch with BugSeq before data submission for optimal host read filtration.
- Improved classification of contigs by masking AMR genes before alignment against the reference database.
- Sequencing depth calculation along with depth distribution plot is now more granular. Previously, BugSeq used the depth of the de novo assembly contigs to predict sequencing depth of the reference genome. As depth of the de novo assembly was aggregated by contig, this process averaged out variation within each contig. With the update, BugSeq uses reads binned by taxonomy to calculate depth of the reference gneome. This process yields maximal resolution for plotting and investigation.
- Scaffold Nanopore and PacBio assemblies for more contiguous assemblies.
- Improved use of assembly graph for classifying Illumina contigs.
- Selecting NCBI nt database with Illumina data uses nt for all stages of analysis, from taxonomic binning of assembly to classification of individual reads. Previously, Illumina analysis with NCBI nt relied on BugRef (default database) for read-level classification.
- Improved reporting of error messages for invalid FASTQ data. A line number where the error occured is now reported if relevant.
- Output MycoSNP SNP distance matrix for Candida auris in Excel format for easier access.
- Percentage of host reads is now calculated after metagenomic classification and includes both the number of reads identified with the host read filter and the number of reads identified as host in metagenomic classiciation. The host read filter remains in preprocessing to protect privacy and increase accuracy.
- Improved identification of Enterovirus and other viral species by adjusting thresholds to determine known versus novel viruses.
- Leverage a chromosome sequence filter to reduce false positive plasmid detections in bacterial isolates.
Warning
This is a breaking change for those using plasmid detection for outbreak investigation. This change results in “selective depletion of plasmids that have integrated within specific lineages but are also autonomously replicating plasmids elsewhere.” Read more and an example use of this filter by Robertson et al (2023).
Fixed¶
Misassemblies,Mismatches/100kbpandIndels/100kpbcolumns were briefly displayed under Assembly Statistics table on the per-sample reports. These columns were previously hidden and are now restored to hidden by default. They are hidden by default as the assembly is compared against the reference genome, so mismatches are often the result of strain variation. Details of this bug are available in MultiQC PR #2190.- PDF reports contained metagenomic classification plots for each taxonomic rank. This has been fixed to the previous behavior of only.
- Issues classifying novel species or those not found in the reference sequence database. Some bacterial isolates may have been overclassified to the nearest species.
- Read preprocessing was briefly reported on separate lines in the general statistics table.
- Bacterial isolate assembly cleaning (removing contigs thought to be from extraneous DNA based on coverage, taxonomic classification and assembly graph connections) was inappropriately applied to some users’ analyses. This may have resulted in removal of contigs from the assembly (less than 1% of total assembly length) containing important genes such as those predicting AMR. Analyses such as refMLST and MLST were not impacted.
- Improved taxonomic identification of C. botulinum and related species (e.g. C. sporogenes) via curation of this section of the database to conform to Brunt et al taxonomy.
5.1 - Oct 20, 2023¶
Added¶
- Annotation of variants with drug fold change in CMV antiviral drug resistance analysis.
- Complete Illumina wastewater analysis, including SARS-CoV-2 variant detection, visualization and aggregation of lab data over time.
- Hepatitis B Virus nanopore analysis, including genotyping, variant calling and variant annotation.
- M. tuberculosis lineage typing.
- Improved quality control of pathogen-specific analyses such as Legionella serogrouping, H. influenzae serotyping, N. meningitidis serotyping, S. pyogenes emm typing, K. pneumoniae typing and more.
- MLST reporting in summary report.
Changed¶
- Completely new AMR prediction analysis. This overcomes several limitations and bugs in the custom ResFinder method previously used, as detailed in v5.0. Both precision and recall are improved across all bacteria, with particular improvements to novel allele detection. This update also brings many new species-level AMR models. A benchmark paper will be published on the new AMR analysis and database; we are currently seeking academic partners interested in validation so reach out if interested.
- Data submitted as bacterial isolates but found to have low level abundance of additional microbes will lead to masking of these additional organisms from reports in order to clarify reporting.
- Improved low complexity filtering for Illumina reads, which leads to more precise taxonomic classification.
- Pathogen identification from metagenomic samples is now reported with a Likert scale of probability from “Very likely” to “Very unlikely”.
- Metagenomic and taxonomic database update.
- Improved deduplication of sequences in the metagenomic database, improving classification accuracy.
- Use of WHO and additional databases for Mycobacterium tuberculosis resistance prediction. Confidence score for TB AMR prediction is now based on WHO confidence. The database source of each variant is annotated in the sample report.
Fixed¶
- Bacterial isolate summaries may have failed to be generated in some bacterial isolate analyses.
5.0 - May 29, 2023¶
Known Issues¶
- AMR gene alleles may be incorrectly called if they have an abundance of silent mutations. The AMR gene family should still be correctly called.
- Multiple AMR genes may be called for the same genomic region/sequence if there is a tie to two or more nearest alleles for that sequence.
- AMR genes reported on plasmids may be discordant from AMR genes reported in the bacterial genome. Plasmid AMR gene detection currently relies on a different method from bacterial genome AMR gene detection, and these methods will be harmonized in a future update.
- Mean Phred scores per base in summary report are calculated as a simple average of all base Phred scores, instead of incorporating considerations for a logarithmic scale. Additional details are available here.
Added¶
- Barcode crosstalk correction for nanopore assembly-based metagenomic abundance calculation.
Warning
This change only affects abundance calculation and does not affect assembly or taxonomic binning. Metagenomic bins for taxa deriving from crosstalk, even if corrected to 0% abundance, will still be output. A future update will hide metagenomic bins purely deriving from barcode crosstalk.
- Name of reference database used to all reports.
- Additional QC metrics included in bacterial isolate summary report, including mean read qualities, assembly N50, assembly L50, percent assembly duplication, and percent identity to reference genome.
- Ability to opt out of outbreak analysis (which adds samples to permanent laboratory database). A check box is now available on the submission page to use this feature.
- Haemophilus influenzae and Neisseria meningitidis serotype prediction.
- Streptococcus pyogenes (Group A Streptococcus) emm typing.
- Reporting of localization of AMR genes to plasmids. See Known Issues for limitations around this reporting.
- Reporting of additional information on detected plasmids, including coverage, and replicon, relaxase and mate-pair formation typing.
- Reporting of percentage of input reads filtered with host read scrubber. This data is located in the general statistics table of the summary report.
- Some results from the per-sample reports are now available in aggregate form in the summary report. This includes:
- Streptococcus pyogenes emm typing
Fixed¶
- The median base quality, both within and across reads, is properly calculated in the summary report. Additional details are available here.
Warning
Median read Q score in the General Statistics was temporarilty reported erroneously based on a median of all reads’ median error probability, instead of a median of all reads’ average error probability. See this GitHub issue for an example. This error tended to inflate the median read Q score statistic.
- Median read length in the General Statistics table of summary report is properly calculated.
- Zip file of summary results may have included a zip file of summary results.
- Capitalization of some taxon names.
- Clarified text in reports describing plasmid cluster IDs.
- Number of reads reported for Illumina analyses in general statistics table of summary report now accurately reflects the number of all reads passing quality control filters. Read counts may have been underestimated before this fix, as only a subset of reads were counted.
- Text now wraps inside cells of tables on per-sample and summary reports.
- Improved detection of AMR predictors from R10.4.1 nanopore sequencing data basecalled with the fast model.
Changed¶
- Assembly completeness in the bacterial isolate summary now reflects both single-copy and duplicated single-copy orthologs. The previous metric reflected only single-copy orthologs, and can be derived from the current metrics as
Assembly Completeness - Assembly Duplication = Unduplicated Assembly Completeness. For pure bacterial isolates, the difference should be less than 3% to Assembly Completeness. - Removed minimizer duplication heatmap from summary reports for Illumina read-based metagenomic classification. This plot did not add value to assessing false positive classifications. Future updates will integrate genome coverage metrics into Illumina read-based metagenomic classification for improved classification precision.
- Stringent demultiplexing is now enabled for nanopore sequencing data by default if submitted without barcoding data or if submitted as FAST5 files.
Note
Files submitted with barcode information in the filename or folder name (if a folder was uploaded) will be unaffected.
- Improved coloring of “Genome Completeness” visualizations to reflect the severity of missing, fragmented and duplicated single-copy orthologs.
- Speed optimizations for large Illumina metagenomic samples. Results should not be affected.
- Major AMR database updates. Improvements are made to multiple beta-lactamase groups, along with better consistency across drugs within the same class.
- Additional selective reporting of antimicrobials for specific taxa (eg. Enterobacter and Serratia spp.).
- New 16S analysis for Illumina sequencing data. Individual reads are error-corrected and classified against a reference database. This new analysis brings increased classification recall and precision on internal benchmarks. For users interested in diversity estimation, we advise to filter taxa with less than 0.1-1% abundance, or to analyse at the genus rank.
- Updated MLST database.
- Updated SARS-CoV-2 lineage database.
- Updated plasmid database for improved plasmid host prediction.
- New default reference sequence database. This update brings significant accuracy improvements to classification of sequences deriving from plasmids.
- Better visualization of detected genotypic markers in the summary report via heatmap-style table.
- Plot sequencing depth by amplicon for amplicon sequencing experiments.
- Novel plasmids are named by the closest sequence in NCBI. The novel plasmid name format is “Novel_
-like” plasmid. - Better precision for Illumina read-level classification.
- Improved host read filtering by building a more robust host reference database.
- Improved host read filterieng for Illumina sequencing data, resulting in improved performance for bacterial isolates with regions of sequence similarity to host organism (eg. Neisseria gonorrhoeae).
- Improved quality control reporting for Salmonella serotyping by including descriptive warnings.
- Improved read quality control for nanopore datasets which were not multiplexed.
- Improved circularity detection for sequences in Illumina sequencing data.
- Remove adapter detection plot in summary report for non-Illumina data.
- Improved assembly of nanopore data by factoring in data characteristics into assembly process and optimizing assembler parameters. Assemblies should be equivalent or better (more contiguous with greater accuracy) across basecaller presets and flowcell chemistries.
- Faster Illumina assembly, with increased contiguity and accuracy from internal benchmarks. The new assembly process also results in reduced gene duplication in taxonomic bins.
4.0 - November 7, 2022¶
Added¶
- QuAISAR-style coverage calculation available in the bacterial isolate summary spreadsheet.
- Conditional formatting of outbreak analysis distance matrices.
- Filter host and Phi X reads before Illumina assembly.
- Monkeypox consensus sequence generation and clade classification.
- Visualization of nanopore and Illumina read-level metagenomic classification results in summary reports.
Note
Illumina read-level metagenomic classification is currently experimental and functionality may change in the future.
- Barcode crosstalk correction for nanopore sequencing data. We follow our previously published algorithm described and validated by Gauthier et al. (2021).
- Report percentage of host reads in the general statistics table of the summary report for nanopore data.
Fixed¶
- BugSeq evaluated a new algorithm to generate our taxonomic classification database. After hearing from our users, this algorithm did not generate databases with the hoped performance and we have reverted the database change.
Warning
Analyses run between August 15 and September 6 may have been affected, and we encourage users to resubmit their data if analysed during this period.
- Sort columns by sample name in summary AMR table.
- Erroneous blank line in some tables of PDF reports.
- Fixed cefixime reporting for ESBLs. Cefixime should now be flagged as having a genotypic predictor of resistance if an ESBL is present.
- Fixed ceftriaxone reporting for carbapenemases. Ceftriaxone should now be flagged as having a genotypic predictor of resistance if certain carbapenemases are present.
- Aggregate plasmid table reported a plasmid with name “0” as found when no plasmids were found.
- Barcode trimming is skipped if reads have already had barcodes trimmed.
- Bacterial isolate summary table is now sorted by sample name.
Changed¶
- All databases were updated. New database include broader taxonomic representation and should therefore provide increased classification accuracy.
- Outbreak analysis module can now handle isolates that have been submitted to BugSeq multiple times with the same name. Each time an isolate is submitted to BugSeq, we now record the date of submission to keep track of resequenced/duplicate isolates.
- Updated AMR database. Major updates are made to OXA-type beta-lactamases, which should more accurately represent the phenotype of individual families and alleles.
- Read count plot has been merged with read filtering plot in summary reports to reduce redundancy of results.
- Nanopore 16S analysis now accepts clusters of 30 reads or larger.
- Stringent demultiplexing for nanopore has temporarily been removed. BugSeq now relies upon the demultiplexing performed by the user. If you would like to analyse stringently demultiplexed data, please perform this before submitting to BugSeq.
- Speed optimizations.
- Keep reads with a greater number of Ns to discard less data.
- Improve nanopore RNA assembly speed and quality.
- Report median read length instead of mean read length in reports.
- Dynamically set contig length suffix (eg. bp, Kbp, Mbp) in reports.
- Filter host (eg. human) reads before metagenomic classification. This improves both speed (fewer reads need to be classified against the full database) and accuracy (some reads may have erroneously been classified to organisms with similar genomes to host).
3.0 - May 16, 2022¶
Added¶
- Flowcell/sequencing run quality control for nanopore sequencing data. If you submit FASTQ files to BugSeq containing sequencing information in the FASTQ headers, this will now be plotted by run ID on the summary report.
- Legionella serogroup prediction.
- Information on read filtering during preprocessing to summary reports for all nanopore sequencing experiments.
- Sample reports now contain additional strain typing information on Salmonella, Klebsiella and Legionella species.
- Bacterial isolate summary report in Excel format.
Fixed¶
- Better detection of nanopore 16S experiments by lowering the acceptable median length of 16S reads.
- Plot titles in reports now reflect the content of the plot instead of the tool used to generate them (which was sometimes erroneous).
- Fixed fraction of referenge genome covered calculation for assemblies which were close to 95% sequence identity to the reference genome. In this scenario, reference genome coverage was vastly underestimated. Bacterial isolates with >99% sequence identity to the reference genome were unlikely to be affected. This issue is related to this bug in QUAST and BugSeq has implemented an internal fix pending an official fix on QUAST.
- refMLST allele calculation if there was a variant in the first or last base of a loci. Accounting for these variants increases resolutions and distance between isolates by an average of 2 alleles at distances less than 50.
- refMSLT clustering of isolates that are equidistant to two separate clusters. Isolates meeting this criteria now cluster with the first cluster observed. Previously, their clustering was assigned randomly to one of the two or more equidistant clusters.
- Platform-specific thresholds for classifying sequencing quality control data as pass/warning/fail.
- Missing tables in PDF reports if they have too many columns. That is now fixed with a message to check the HTML report for the full table.
Changed¶
- Summary reports now show total reads after filtering. This streamlines the summary reports for Illumina paired-end data as individual FASTQ files are no longer reported in the General Statistics table by default. Note that the number of reads after filtering is the sum of both paired-end files for Illumina.
- Krona plots for BugSplit (assembly-level) classification now shows all ranks, including intermediary ranks.
- Unclassified contigs (and the reads mapping to them) are now classified as unclassified instead of root in Kraken report formats.
- No longer expose unbinned assemblies. The unbinned assembly may be obtained by concatenating together all of the binned assemblies.
- Removal of Racon/Medaka/Homopolish polishing of nanopore R9.4.1 and R10.3 assemblies for the following reasons:
- Unfortunately, both Racon and Homopolish had critical issues preventing their widespread use.
- On internal benchmarks, Medaka may decrease assembly quality if there is a mismatch between basecaller version and Medaka model.
- With recent improvements in nanopore basecalling accuracy, polishing no longer has a significant impact on assembly quality.
- Polishing was adding to processing time but was not increasing metagenomic classification or downstream analysis accuracy (eg. AMR). BUSCO completion analysis may be impacted with more fragmented genes detected; however, this does not impact other analyses.
- Faster Illumina analyses by allocating more CPUs to the assembly process.
- Do not bin long-read assembled contigs less than 1000bp in length. This increases accuracy of analysis as these contigs were the most likely to be erroneously classified, and should also not be present in long-read assemblies.
- Expose additional antibiotics on sample reports for Mycobacterium tuberculosis.
- Allow IUPAC characters in input sequencing data.
- Improved taxonomic binning of bacterial isolates.
- Updated AMR and MLST database.
- Bin plasmids to their host bacteria across all sample types.
2.3 - January 25, 2022¶
Added¶
- Include the following data in individual sample reports:
- Antimicrobial resistance table
- TB spoligotyping
- Sample name
- Table of plasmids detected
- Canonical SNP typing for:
- B. anthracis
- F. tularensis
- Y. pestis
- C. burnetii
- Include the followind data in aggregate reports:
- Plasmids detected across all samples
- Antimicrobial resistance detection across all samples
- Read filtering statistics
- PDF generation of reports
- Assembly and processing of Q20+ ONT sequencing data
- Insertion and deletion detection for Bacillus anthracis
- cDNA amplification primer trimming on metatranscriptomic nanopore data
Fixed¶
- Error handling if a user submits duplicated reads across multiple files
- Coverage per contig plot in sample reports
- Reporting of first line antimicrobials for Mycobacterium tuberculosis
Changed¶
- Improved detection of plasmid sequences
- Improved assembly of ONT cDNA/RNA-sequencing data
- Improved abundance calculation from Illumina assemblies
- Improved primer scheme detection and trimming for SARS-CoV-2 amplicon sequencing data
- Faster nanopore 16S processing by setting the QIIME2 vsearch
--maxrejectsflag. On internal evaluations, this has neglibile impact on results but drastically speeds up processing. - Faster overall analyses by being more selective in which pathogen-specific analyses get run
- Gentler quality filtering for Illumina data should result in better and more contiguous assemblies
- Faster read preprocessing by better leveraging parallel processing
- Removed raw text file output of AMR data as it is now contained in reports and much easier to use
2.2 - October 26, 2021¶
Added¶
- Plot BUSCO results in reports
- Include MLST results in per-sample report
- Report analysis pipeline version in reports
Fixed¶
- Bug handling many Illumina samples in one analysis
- Depth calculations only included reference contigs with mapping assembled contigs. This is now fixed - sequencing depth may be slightly lower than previously reported.
Changed¶
- BugSplit can now report strain-level classifications
- Upgraded
seqkitfor faster FASTQ parsing
2.1 - October 14, 2021¶
Added¶
- Per-sample HTML report
- Sequencing depth calculation performed for each genome in a sample relative to its reference sequence
- Stringent demultiplexing of user submissions with custom file names
- Visualization of Pangolin SARS-CoV-2 lineage results in the summary report
Fixed¶
Changed¶
- Improved taxonomic binning of Illumina sequences using assembly graph information
- Improved Illumina assembly by merging paired-end reads
- Faster internal file transfers, resulting in faster analyses
2.0 - October 7, 2021¶
Added¶
- Assembly and polishing of data from all sequencers
- BugSplit module: high-accuracy taxonomic binning (Citation)
Fixed¶
- Respect a user’s custom filename
Changed¶
- Only demultiplex nanopore if it does not have a custom filename
1.0.0 - December 3, 2019¶
Added¶
- First release!