Metadata submission¶
Metadata can be submitted alongside sequencing data and is incorporated into various parts of our platform. Examples of annotating samples and the subsequent impact include:
- Samples can be specified as a negative control. Read counts in other samples will be normalized against the read counts in the negative control.
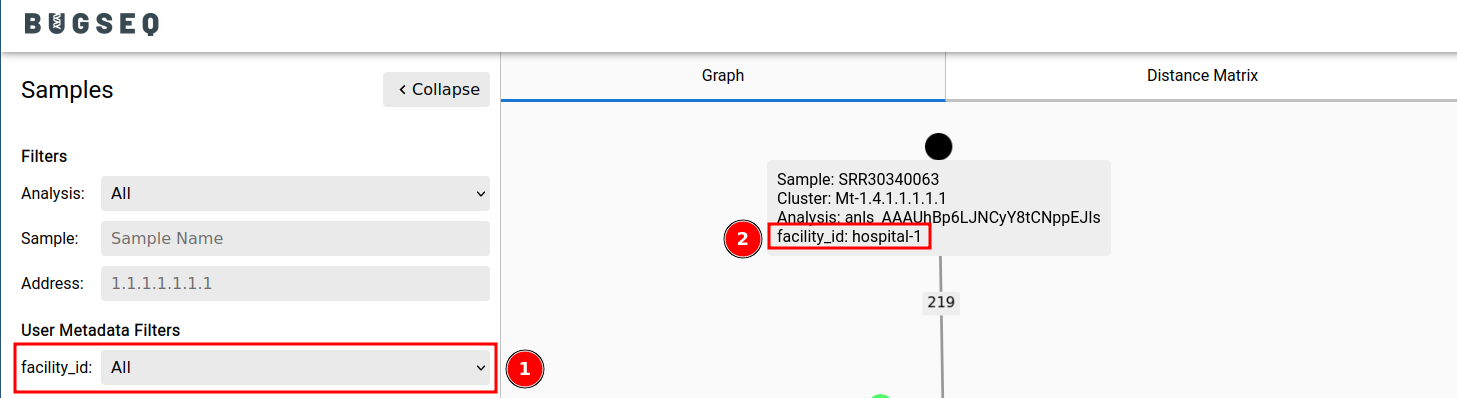
- Samples can be annotated with arbitrary fields. For example, a sample can be annotated with
facility_id: hospital-1. The metadata will appear in outbreak analyses.
Specifying metadata keys for a lab¶
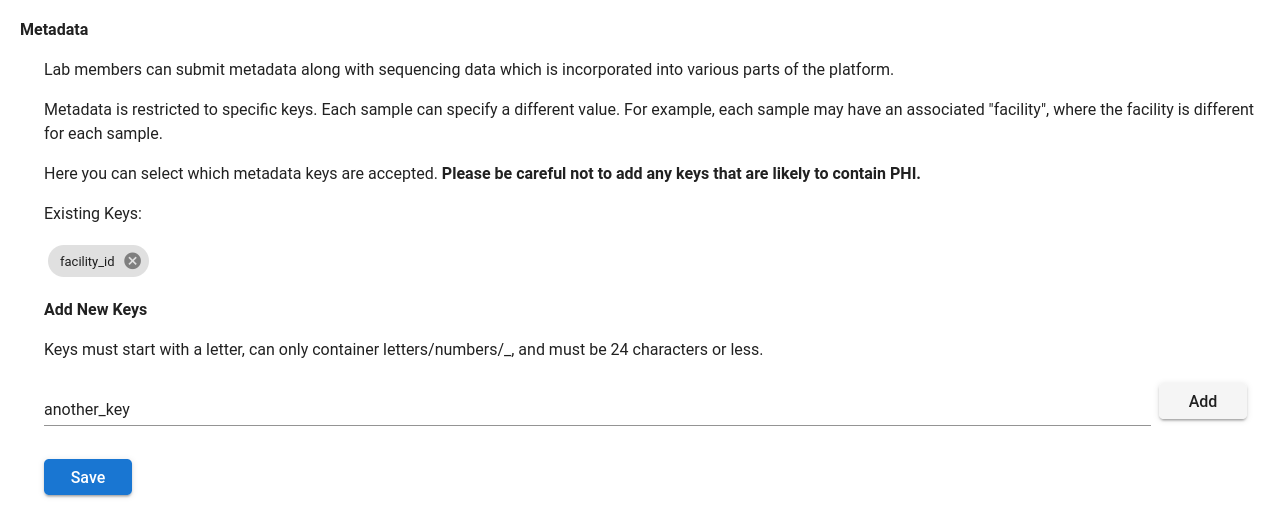
To annotate a sample with metadata, a lab manager must add the key to the lab as an eligible metadata key. Metadata keys can be configured under “Labs”, on the “Data Access & Notifications” tab. Here you can add or remove keys.
Please be careful not to add any keys that are likely to contain protected health information.
Annotating a sample with metadata¶
We recommend uploading files first, as the upload form will help annotate recognized samples.
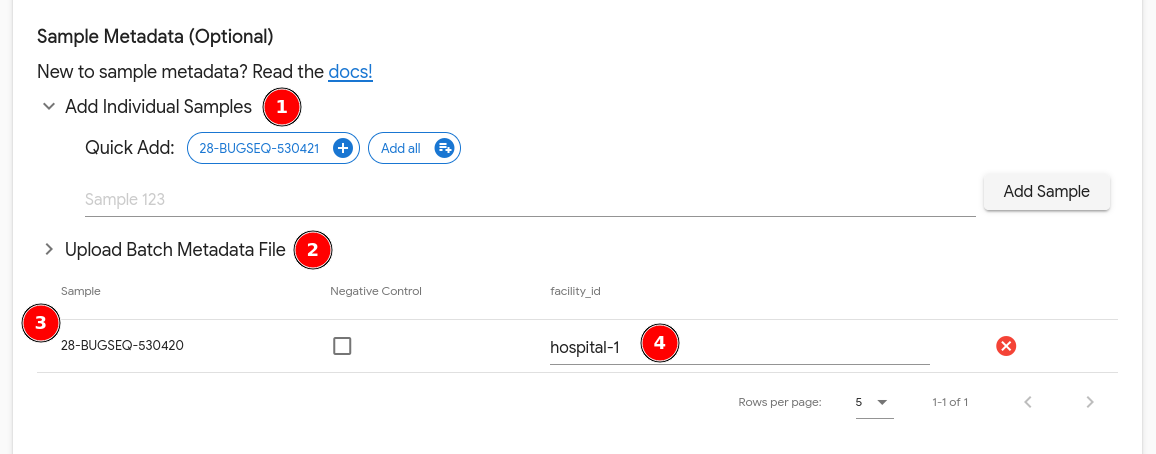
Samples can be added in 2 ways:
- Add each sample individually. If sample names can be detected from input files, click on the “Quick Add” samples to add them directly. Otherwise you can manually input a sample name and click “Add Sample”.
-
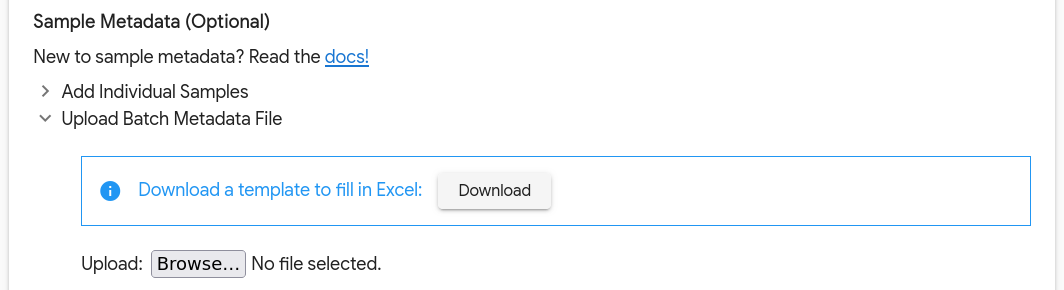
Add samples in bulk using a CSV file. The permitted columns are
sample(the name of the sample),negative_control(values can betrueorfalse) and any custom metadata keys enabled on the lab that the analysis is being submitted under. We recommend downloading the provided template and filling it out in Excel.
Not all submitted samples need to be annotated with metadata.
Warning
All samples that are annotated must have matching sequencing data submitted. If metadata is submitted for a sample that doesn’t have any sequencing data, the analysis will fail.