Labs¶
BugSeq strives to empower labs with many users to collaborate, share and aggregate data across members and analyses.
Some features we offer for labs:
- Sharing analyses with some or all members of a lab (labs can be set up to automatically share all analyses)
- Aggregating outbreak information from all analyses submitted within a lab - see outbreak analysis
- Querying for genomes seen by other members of the lab, and tying those back to specific samples (coming soon)
- Stay tuned for more features!
Creating a lab¶
One user must create the lab. They will be the designated lab manager.
-
Head over to the Labs section on the left navigation bar:
-
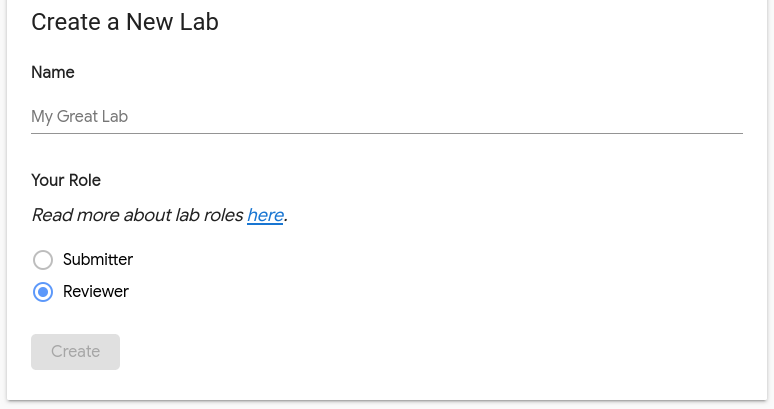
Enter a name for the lab, select a role and click Create
Note
For more about roles, see the roles documentation.
Inviting members¶
Once you have a lab set up, you can invite your lab members.
-
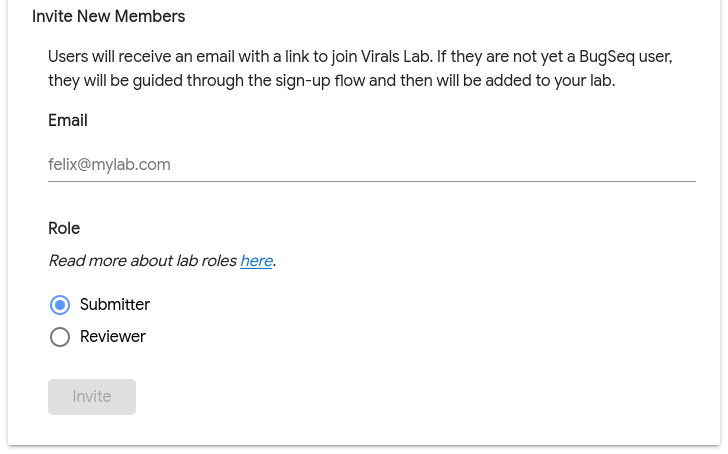
On the Labs page, there will be a section to Invite New Members:
-
Fill in an email address, select a role and click Invite
Note
For more about roles, see the roles documentation.
Note
The invited user doesn’t yet need to have a BugSeq account, and the email entered doesn’t need to match the email that they use for their BugSeq account.
-
An email will be sent to the user with instructions to join the lab. If they haven’t yet registered for BugSeq, it will guide them through that flow and then add them to the lab.
Lab settings¶
Analysis parameters¶
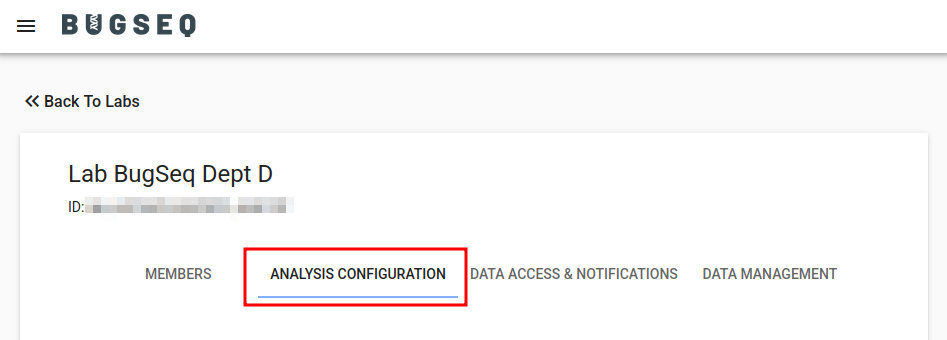
Per-lab analysis configuration can be configured on the Labs page under “Analysis Configuration”
Pipeline parameters¶
Pipeline parameters control thresholds where alerts and warnings will appear in executive summaries on per-sample and summary reports. For example, here are a few options that can be configured:
-
Percent Identity to Reference Genome Pass/Fail ThresholdThis enables you to configure alerts if a bacterial isolate’s sequence identity to the reference genome doesn’t meet this threshold.
-
Assembly Length Outlier Detection Warning ThresholdDetects outliers for assembly length relative to the species’ distribution using Tukey’s fences test.
Cluster alerting¶
Cluster alerting parameters control when the per-sample reports will include alerts about samples being added to ongoing clusters. The sensitivity of alerting is configurable via a default and per-species allele threshold. This enables labs to set a default alert threshold, with overrides for species that are known to mutate faster or slower.
A Default Alert Threshold will apply to any organisms that don’t have their own thresholds specified. These will yield alerts such as:
- Sample was added to existing cluster prefix: '2.1.1.1'
Per-organism alert thresholds can be configured for organisms where the lab is only interested in differences at a certain allele cutoff. For example, labs can configure alerts when a sample is added that’s within 100 alleles of an existing sample analyzed under the same lab.
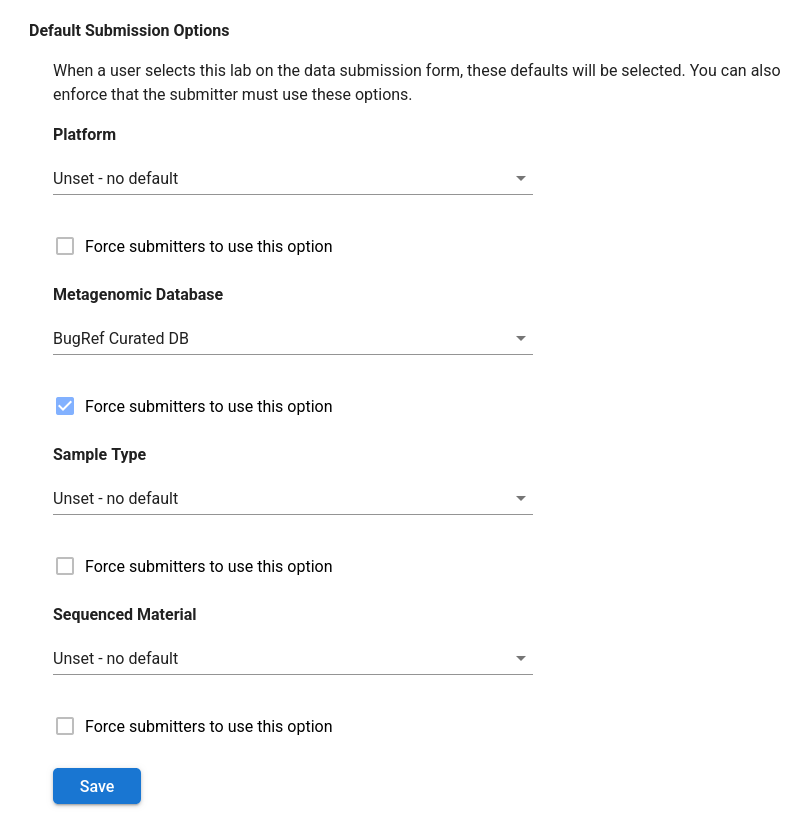
Default submission options¶
You can configure a lab with defaults for the submission options, such as platform and database. This is helpful if you routinely submit the same kind of data to a given lab. Furthermore, you can enforce that all members must submit using a predefined set of options to ensure uniformity within that lab.
Sharing¶
Data sharing can be configured within a lab to meet the privacy requirements of the lab.
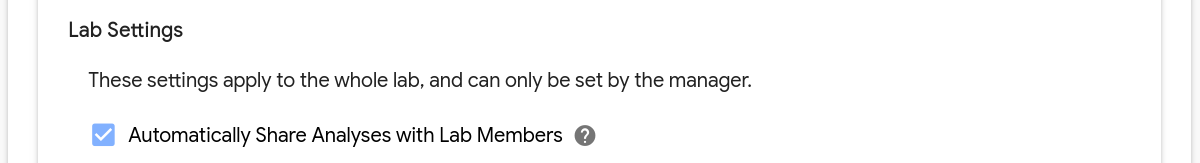
Note the setting to automatically share analyses with lab members:
For labs with this setting enabled, when an analysis completes that was submitted by a member of the lab, all members will be able to access the analysis. Members that haven’t opted out of emails (see individual settings) will also receive an email notification with a link to access results.
When to use
This configuration is great for labs that work tightly together, where all members should be notified of activity.
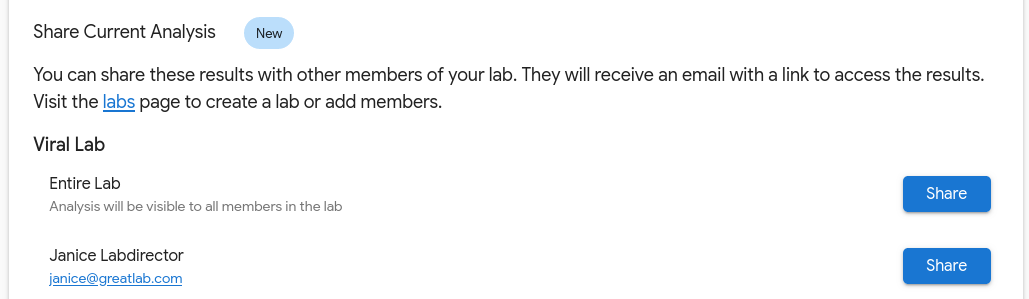
If the setting isn’t enabled, analyses can still be shared. On the analysis results page, you will find the option to manually share the analysis with the entire lab or individual lab members. This mechanism shares a single analysis at a time.
When looking at an analysis, scrolling down will reveal sharing options:
When to use
This configuration is great for labs that analyze high volumes of data and want to flag specific analyses to epidemiologists or lab directors.
Individual settings¶
Users can set individual settings for each lab that they’re a member of.
When “Receive an Email When an Analysis is Shared with the Entire Lab” is checked, you will be notified when an analysis is shared with the entire lab. The notification will occur when an analysis completes if “Automatically Share Analyses with Lab Members” is enabled, or if the analysis owner manually shares it with the entire lab on the results page.
You will be notified if a lab member directly shares an analysis with you - this isn’t configurable.
Roles¶
A lab member’s role controls the options and data that are available to them.
A lab member’s role is determined when they’re invited to the lab.
All lab members will show up as part of a lab. If the lab has automated sharing enabled, all members will gain access to new analyses as they complete. An analysis owner can also manually choose to share analyses with any lab member of any lab the analysis owner is a member of.
The two main roles are:
-
Submitter
A Submitter can submit data for analysis. This is an ideal role for most members of most labs.
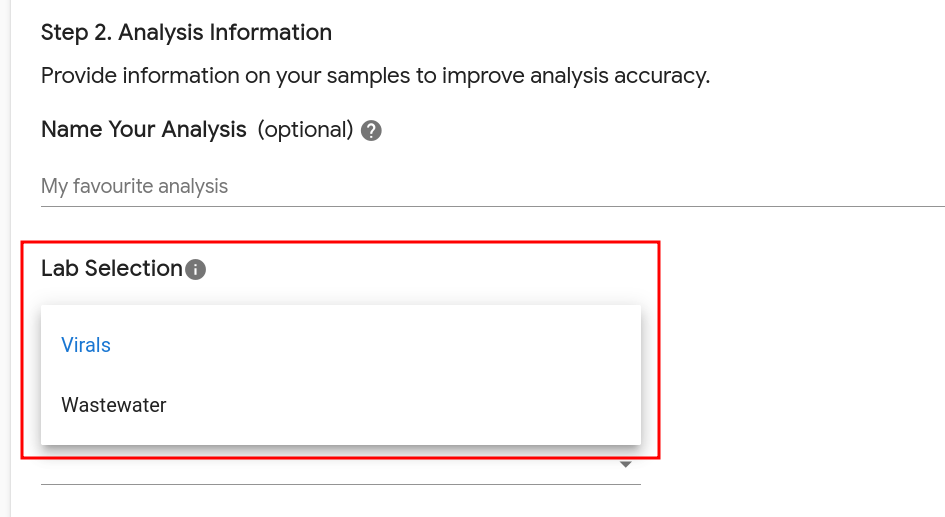
Note that if a user is a Submitter in multiple labs, they will need to select which lab’s data repository to use when submitting data:
This informs BugSeq of which data repository to use for outbreak analysis, wastewater analysis, and other types of analysis that aggregate data across analyses from the same lab.
Tip: Members should be a submitter of only one lab
We recommend that most users only be a Submitter in one lab. Being a Submitter in one lab means that the member doesn’t need to choose which lab to submit data under when submitting data for analysis. This reduces complexity and possibility of error.
-
Reviewer
A Reviewer can access shared analysis data but can’t submit data for analysis. This is an ideal role for supervisors, epidemiologists and other members that need broad read-only access.